 |
WEB-based GEne SeT AnaLysis Toolkit
|
| |
| Translating gene lists into biological insights... |
| |
Features unique to WebGestalt or shared with other tools.
The purpose of this table is to distinguish which WebGestalt features are unique to WebGestalt and which of them are available in other tools. Thus, the table only lists features available in WebGestalt. This is not meant to be a comprehensive comparison of enrichment analysis tools, and only three other representative tools are included:
DAVID, g:Profiler and FatiGO.
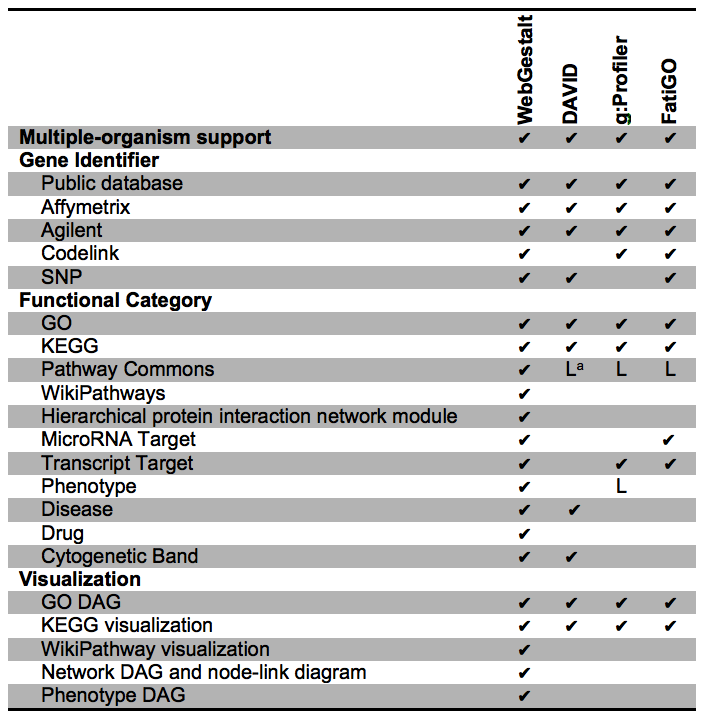
a:DAVID and FatiGO contains Reactome and Biocarta databases while g:Profiler contains Reactome and BIOGRID databases. These databases are a portion of the Pathway Commons database; g:Profiler only contains phenotypes from Human Phenotype Ontology.
WebGestalt is currently developed and maintained by Jing Wang and Bing Zhang at the Zhang Lab. Other people who have made significant contribution to the project include Dexter Duncan, Stefan Kirov, Zhiao Shi, and Jay Snoddy.
Funding credits: NIH/NIAAA (U01 AA016662, U01 AA013512); NIH/NIDA (P01 DA015027); NIH/NIMH (P50 MH078028, P50 MH096972); NIH/NCI (U24 CA159988); NIH/NIGMS (R01 GM088822).


